Reconstruction and Registration of Histology and Phase Contrast Images for Clinical Validation of Imaging ModalitiesIn medical collaboration with:Dr. Christian Glaser, Silvia Adam-Neumair In academic collaboration with: Prof. Dr. Franz Pfeiffer, Dr. Paola Coan, Prof. Dietrich Habs This project is funded by the Munich Center of Advanced Photonics (MAP). http://www.munich-photonics.de/ Scientific Director: Prof. Dr. Nassir Navab Contact Person(s): Martin Groher |
Abstract
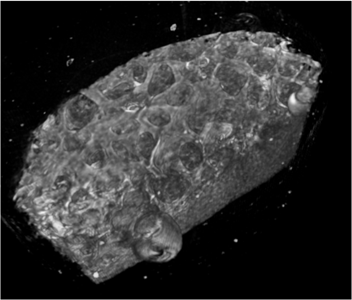
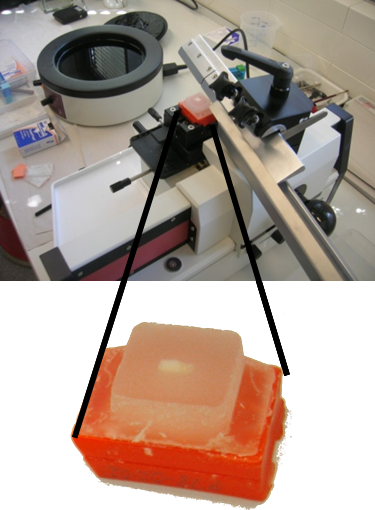
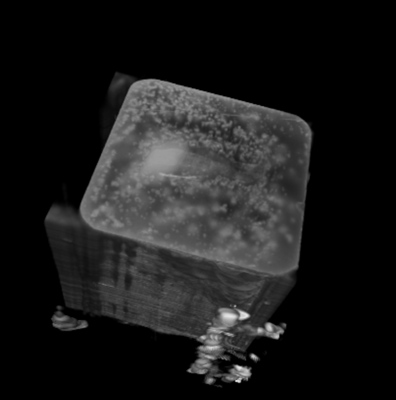
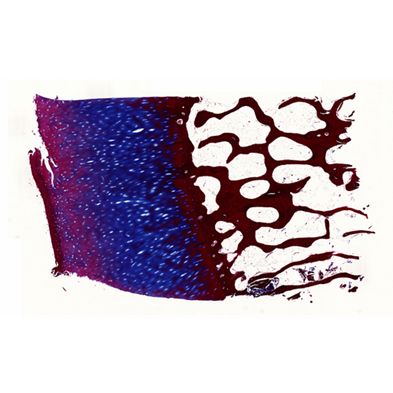
Before its introduction into the hospital, a new imaging modality has to be validated. In other words, appearing structures need to be correlated to the imaged tissues. Such a cross validation is only meaningful when performed against the gold standard which is histology. Currently, cross validation is performed by qualitative comparison of 3D datasets to 2D histology slices. Since the acquisition of a consistent 3D histology volume is a challenging task, its comparison to the corresponding dataset always remained qualitative.The classic histology procedure can be divided in four steps: pre processing, cutting, post processing and imaging of the tissues. In the first step, the sample is chemically processed to preserve the tissues and is then embedded in a paraffin block. By using a microtome, it is cut in very thin slices, and put on a glass slide. During post processing the sample is stained to enhance the structures of interest. The imaging is performed with a camera mounted in a microscope, or with a dedicated scanner. Several difficulties inherent to this process can have a dramatic influence on the quality of the reconstructed histology volume. For instance, since the cutting process is done manually, problems like flipping, bending or ripping of the slides may happen. If the knife starts to wear off, some banding will appear over the slices. Moreover, a few slices could be missing. Finally since the staining color is time dependent, variation in the color of the slices can occur.
In this project, we propose to improve the histology procedure and to develop methods towards a consistent reconstruction of 3D histology volumes.
Detailed Project Description
Pictures
|
|
|
|
Publications
| 2012 | |
| Y. Yagi, M. Groher, M. Feuerstein, M. Onozato, H. Heibel, N. Navab
Overcoming Challenges in Histology 3D Imaging 11th European Congress on Telepathology and 5th International Congress on Virtual Microscopy, Venice, Italy, June 2011. (bib) |
|
| 2011 | |
| M. Feuerstein, H. Heibel, J. Gardiazabal, N. Navab, M. Groher
Reconstruction of 3-D Histology Images by Simultaneous Deformable Registration Proceedings of Medical Image Computing and Computer-Assisted Intervention (MICCAI 2011), Toronto, Canada, September 2011. The original publication is available online at www.springerlink.com (bib) |
|
Team
Contact Person(s)
|
Working Group
|
|
|
|
|
|
Location
| Technische Universität München Institut für Informatik / I16 Boltzmannstr. 3 85748 Garching bei München Tel.: +49 89 289-17058 Fax: +49 89 289-17059 |
| Ludwig-Maximilians-Universität München Chirurgische Klinik und Poliklinik - Innenstadt, AG CAS Nussbaumstr. 20 80336 München Room: C2.06 Tel.: +49 89 5160-3615 Tel.: +49 89 5160-3635 Fax: +49 89 5160-3630 |
internal project page
Please contact Martin Groher for available student projects within this research project.