IFL Projects
|
|
Learn how to successfully identify unmet clinical needs within the clinical routine and work towards possible and realistic solutions to solve those needs. Students will get to know tools helping them to be successful innovators in medical technology. This will include all steps from needs finding and selection to defining appropriate solution concepts, including the development of first prototypes. Get introduced to necessary steps for successful idea and concept creation and realize your project in an interdisciplinary teams comprising of physicists, informations scientists and business majors. During the project phase, you are supported by coaches from both industry and medicine, in order to allow for direct and continuous exchange.
|
|
|
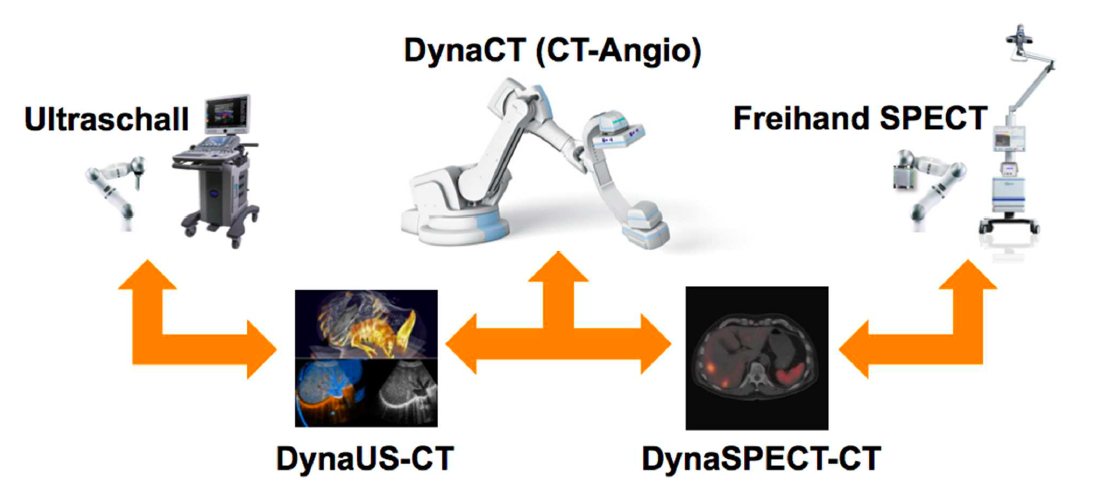
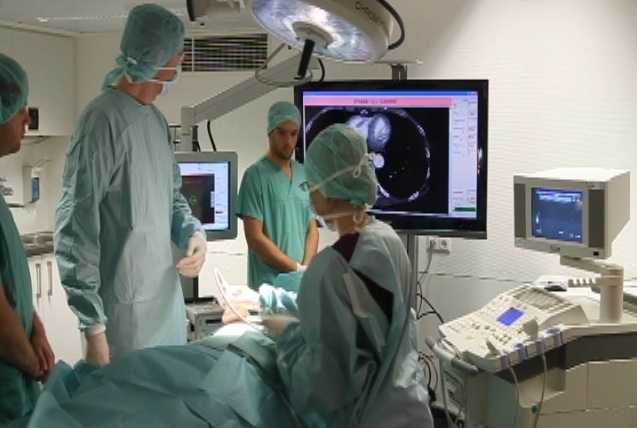
This project aims at developing advanced methods for robotic image acquisitions, enabling more flexible, patient- and process-specific functional and anatomical imaging with the operating theater. Using robotic manipulations, co-registered and dynamic imaging can be provided to the surgeon, allowing for optimal implementation of preoperative planning. In particular, this projects is the first one developing concepts for intraoperative SPECT-CT, and introduces intraoperative robotic Ultrasound imaging based on CT trajectory planning, enabling registration with angiographic data. With distinguished partners from Bavarian industry, this project has a fundamental contribution in developing safe, reliable, flexible, and multi-modal imaging technologies for the operating room of the future.
|
|
|
3D ultrasound imaging has high potential for various clinical applications, but often suffers from high operator-dependency and the directionality of the acquired data. State-of-the-art systems mostly perform compounding of the image data prior to further processing and visualization, resulting in 3D volumes of scalar intensities. This work presents computational sonography as a novel concept to represent 3D ultrasound as tensor instead of scalar fields, mapping a full and arbitrary 3D acquisition to the reconstructed data. The proposed representation compactly preserves significantly more information about the anatomy-specific and direction-depend acquisition, facilitating both targeted data processing and improved visualization. We show the potential of this paradigm on ultrasound phantom data as well as on clinically acquired data for acquisitions of the femoral, brachial and antebrachial bone. Further investigation will consider additional compact directional-dependent representations on the one hand and on the other hand modify Computational Sonography from working on B-Mode images to RF-envelope statistics, motivated by the statistical process of image formation. We will show the advantages of the proposed improvements on simulated ultrasound data, phantom and clinically acquired ultrasound data.
|
|
|
EDEN2020 (Enhanced Delivery Ecosystem for Neurosurgery) aims to develop the gold standard for one-stop diagnosis and treatment of brain disease by delivering an integrated technology platform for minimally invasive neurosurgery. A team of first-class industrial partners (Renishaw plc. and XoGraph ltd.), leading clinical oncological neurosurgery team (Università di Milano, San Raffaele and Politecnico di Milano) lead by Prof. Lorenzo Bello and the involvement of leading experts in shape sensing (Universitair Medisch Centrum Groningen) under supervision of Prof. Dr. Sarthak Misra The project is coordinated by Dr. Rodriguez y Baena, Imperial College London. His team provides the core technology for the envisioned system, the bendable robotic needle. During the course of EDEN2020 this interdisciplinary team will work on the integration of 5 key concepts, namely (1) pre-operative MRI and diffusion-MRI imaging, (2) intra-operative ultrasounds, (3) robotic assisted catheter steering, (4) brain diffusion modelling, and (5) a robotics assisted neurosurgical robotic product (the Neuromate), into a pre-commercial prototype which meets the pressing demand for better and less invasive neurosurgery. Our chair will be focusing on the imaging components (i.e. (1) and (2)), targeting the realtime compensation of tissue movement and accurate localization of the flexible catheters at hand. We will further extend the findings of FP7 ACTIVE, in which we successfully combined pre-operative MRI with intra-operative US through deformable 3D-2D registration, making us most qualified for this role.
|
|
|
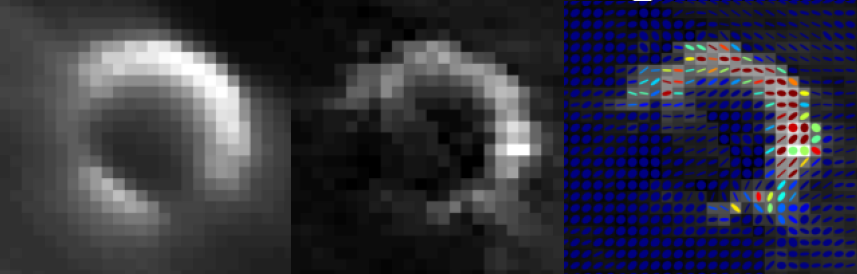
Nuclear medicine imaging modalities assist commonly in surgical guidance given their functional nature. However, when used in the operating room they present limitations. Pre-operative tomographic 3D imaging can only serve as a vague guidance intra-operatively, due to movement, deformation and changes in anatomy since the time of imaging, while standard intra-operative nuclear measurements are limited to 1D or (in some cases) 2D images with no depth information. To resolve this problem we propose the synchronized acquisition of position, orientation and readings of gamma probes intra-operatively to reconstruct a 3D activity volume. In contrast to conventional emission tomography, here, in a first proof-of-concept, the reconstruction succeeds without requiring symmetry in the positions and angles of acquisition, which allows greater flexibility and thus opens doors towards 3D intra-operative nuclear imaging.
|
|
|
Current tracking solutions routinely used in a clinical, potentially surgically sterile, environment are limited to mechanical, electromagnetic or classic optical tracking. Main limitations of these technologies are respectively the size of the arm, the influence of ferromagnetic parts on the magnetic field and the line of sight between the cameras and tracking targets. These drawbacks limit the use of tracking in a clinical environment. The aim of this project is the development of so-called inside-out tracking, where one or more small cameras are fixed on clinical tools or robotic arms to provide tracking, both relative to other tools and static targets.
These developments are funded from the 1st of January 2016 to 31st of December 2017 by the ZIM project Inside-Out Tracking for Medical Applications (IOTMA).
|
|
|
Computerized medical systems play a vital role in the operating room, yet surgeons often face challenges when interacting with these systems during surgery. In this project we are aiming at analyzing and understanding the Operating Room specific aspects which affect the end user experience. Beside operating room specific usability evaluation approaches in this project we also try to improve the preliminary intra-operative user interaction methodologies.
|
|
|
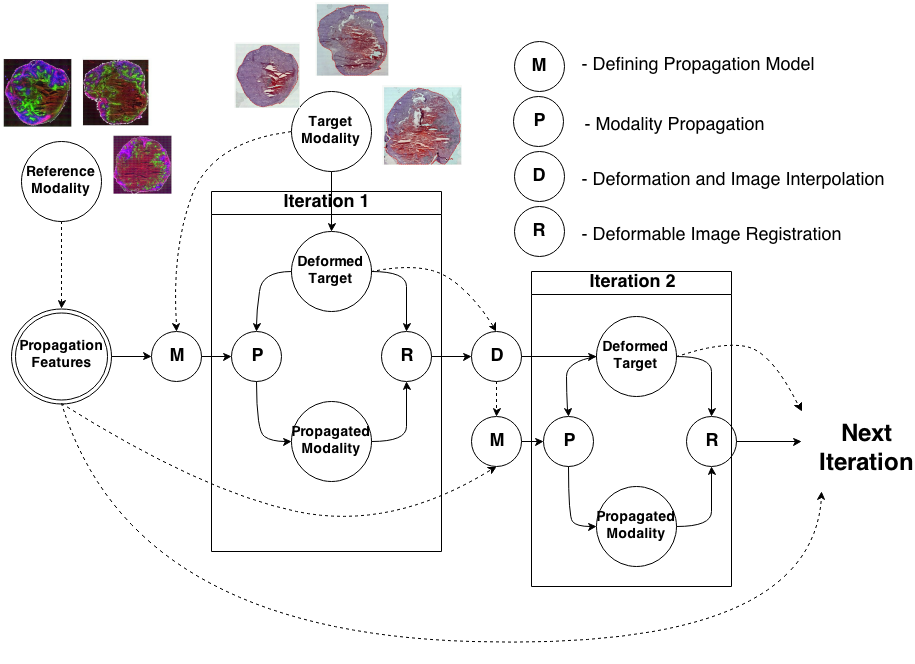
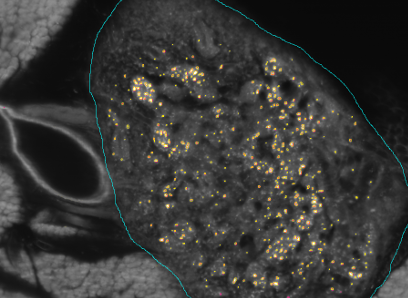
To register modalities with complex intensity relationships, we leverage machine learning algorithm to cast it into a mono modal registration problem. This is done by extracting tissue specific features for propagating anatomical/structural knowledge from one modalitiy to an other through an online learnt propagation model. The registration and propagation steps are iteratively performed and refined. For proof-of-concept, we employ it for registering (1) Immunofluorescence to Histology images and (2) Intravascular Ultrasound to Histology Images.
|
|
|
The SFB824 (Sonderforschungsbereich 824: Central project for histopathology, immunohistochemistry and analytical microscopy) represents an interdisciplinary consortium which aims at the development of novel imaging technologies for the selection and monitoring of cancer therapy as an important support for personalized medicine. Z2, the central unit for comparative morphomolecular pathology and computational validation, provides integration, registration and quantification of data obtained from both macroscopic and (sub-)cellular in-vivo as well as ex-vivo imaging modalities with tissue-based morphomolecular readouts as the basis for the development and establishment of personalized medicine. In order to develop novel imaging technologies, co-annotation and validation of image data acquired by preclinical or diagnostic imaging platforms via tissue based quantitative morphomolecular methods is crucial. Light sheet microscopy will continue to close the gap between 3D data acquired by in-vivo imaging and 2D histological slices especially focusing on tumor vascularization. The Multimodal ImagiNg Data Flow StUdy Lab (MINDFUL) is a central system for data management in preclinical studies developed within SFB824. Continuing the close collaboration of pathology, computer sciences and basic as well as translational researchers from SFB824 will allow the Z2 to develop and subsequently provide a broad variety of registration and analysis tools for joint imaging and tissue based image standardization and quantification.
The goal of the BFS Project: ImmunoProfiling using Neuronal Networks (IPN2) is to develop a method based on neuronal networks and recent advances in Deep Learning to allow characterization of a patient's tumor as ″hot″ or ″cold″ tumor depending on the identified ImmunoProfile. Recent research has shown that many tumors are infiltrated by immuno-competent cells, as well as that the amount, type and location of the infiltrated lymph nodes in primary tumors provide valuable prognostic information. In contrast to a ″cold tumor″, a ″hot tumor″ is characterized by an active immune system which the tumor has identified as threat. This identification provides the basis for selecting the therapy best suitable for the individual patient.
|
|
|
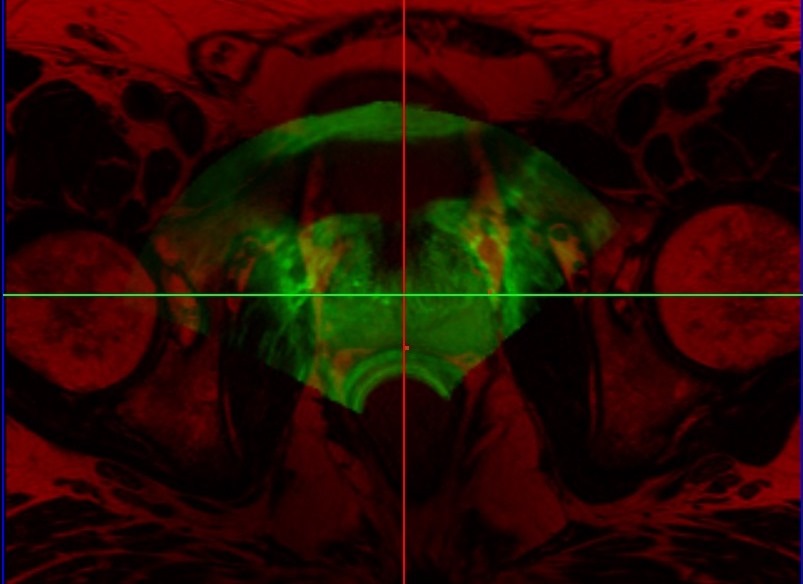
Transrectal ultrasound (TRUS) guided biopsy remains the gold standard for diagnosis. However, it suffers from low sensitivity, leading to an elevated rate of false negative results. On the other hand, the recent advent of PET imaging using a novel dedicated radiotracer, Ga-labelled PSMA (Prostate Specific Membrane Antigen), combined with MR provides improved preinterventional identification of suspicious areas. Thus, MRI/TRUS fusion image-guided biopsy has evolved to be the method of choice to circumvent the limitations of TRUS-only biopsy. We propose a multimodal fusion image-guided biopsy framework that combines PET-MRI images with TRUS. Based on open-source software libraries, it is low cost, simple to use and has minimal overhead in clinical workflow. It is ideal as a research platform for the implementation and rapid bench to bedside translation of new image registration and visualization approaches.
|
|
|
SUPRA is an open-source pipeline for fully software defined ultrasound processing for real-time applications. Covering everything from beamforming to output of B-Mode images, SUPRA can help to improve the reproducibility of results and does allow for a full customization of the image acquisition workflow. Including all processing stages of a common ultrasound pipeline, it can be executed in 2D and 3D on consumer GPUs in real-time. Even on hardware as small as the CUDA enabled Jetson TX2, SUPRA allows for 2D imaging in real-time. You can access the code on our github page: https://github.com/IFL-CAMP/supra Additional information can be found in our work on SUPRA Göbl, R. and Navab, N. and Hennersperger, C., SUPRA: Open Source Software Defined Ultrasound Processing for Real-Time Applications, eprint arXiv:1711.06127, Nov 2017, under review for IPCAI2018 The development of SUPRA was partly funded by the European Horizon 2020 Project EDEN2020.
|