Histology Projects
|
|
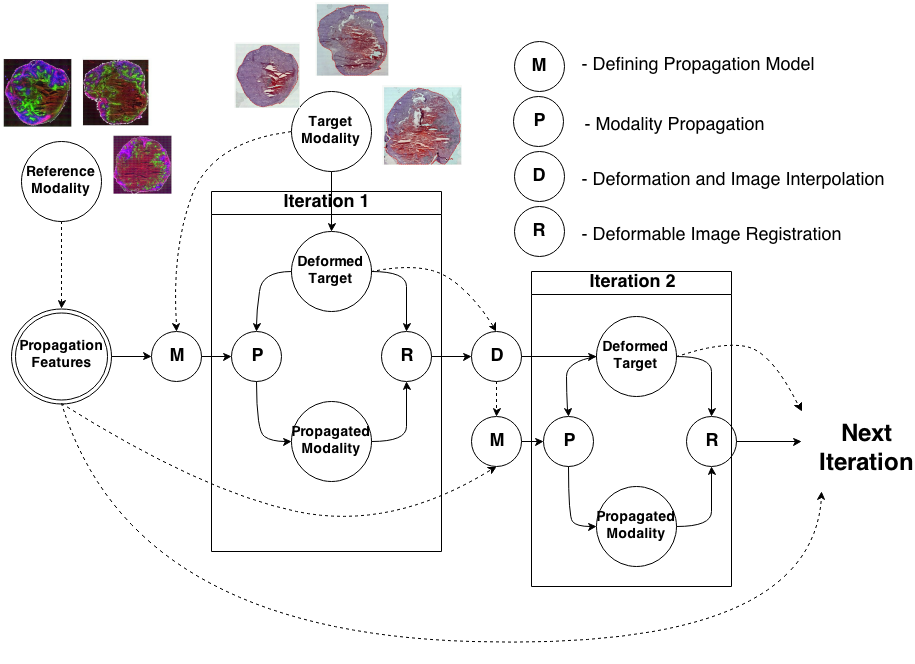
To register modalities with complex intensity relationships, we leverage machine learning algorithm to cast it into a mono modal registration problem. This is done by extracting tissue specific features for propagating anatomical/structural knowledge from one modalitiy to an other through an online learnt propagation model. The registration and propagation steps are iteratively performed and refined. For proof-of-concept, we employ it for registering (1) Immunofluorescence to Histology images and (2) Intravascular Ultrasound to Histology Images.
|
|
|
The SFB824 (Sonderforschungsbereich 824: Central project for histopathology, immunohistochemistry and analytical microscopy) represents an interdisciplinary consortium which aims at the development of novel imaging technologies for the selection and monitoring of cancer therapy as an important support for personalized medicine. Z2, the central unit for comparative morphomolecular pathology and computational validation, provides integration, registration and quantification of data obtained from both macroscopic and (sub-)cellular in-vivo as well as ex-vivo imaging modalities with tissue-based morphomolecular readouts as the basis for the development and establishment of personalized medicine. In order to develop novel imaging technologies, co-annotation and validation of image data acquired by preclinical or diagnostic imaging platforms via tissue based quantitative morphomolecular methods is crucial. Light sheet microscopy will continue to close the gap between 3D data acquired by in-vivo imaging and 2D histological slices especially focusing on tumor vascularization. The Multimodal ImagiNg Data Flow StUdy Lab (MINDFUL) is a central system for data management in preclinical studies developed within SFB824. Continuing the close collaboration of pathology, computer sciences and basic as well as translational researchers from SFB824 will allow the Z2 to develop and subsequently provide a broad variety of registration and analysis tools for joint imaging and tissue based image standardization and quantification.
The goal of the BFS Project: ImmunoProfiling using Neuronal Networks (IPN2) is to develop a method based on neuronal networks and recent advances in Deep Learning to allow characterization of a patient's tumor as ″hot″ or ″cold″ tumor depending on the identified ImmunoProfile. Recent research has shown that many tumors are infiltrated by immuno-competent cells, as well as that the amount, type and location of the infiltrated lymph nodes in primary tumors provide valuable prognostic information. In contrast to a ″cold tumor″, a ″hot tumor″ is characterized by an active immune system which the tumor has identified as threat. This identification provides the basis for selecting the therapy best suitable for the individual patient.
|
|
|
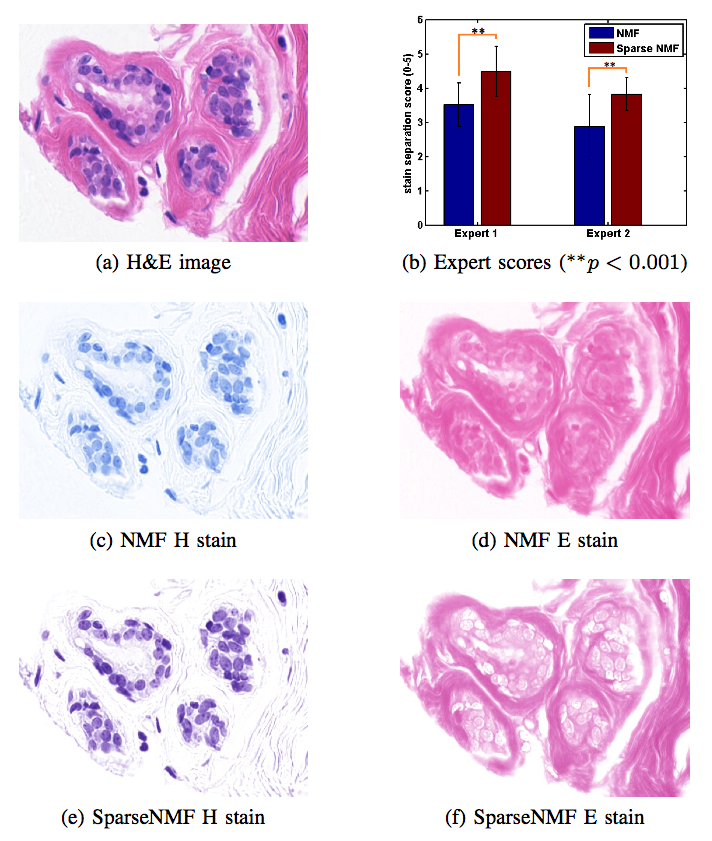
Staining and scanning of tissue samples for microscopic examination is fraught with unwanted variations that affect their color appearance. Sources of these variations include differences in raw material and manufacturing techniques of stain vendors, staining protocols of labs, and color responses of digital scanners. Color normalization of stained biopsies and tissue microarrays will help pathologists and computational pathology software while comparing different tissue samples. However, techniques that are used for natural images, such as histogram matching fail to utilize unique properties of stained tissue samples and produce undesirable artifacts. Tissue samples are stained with only a few reagents (frequently only two -- hemotoxylin and eosin or H\&E) and most tissue regions bind to only one stain or the other, thus producing sparse density maps composed of only a few components. This underlying structure of sparse stain density is biomedically important. We used these properties of stained tissue to propose a technique for stain separation and color normalization. Based on sparse non-negative matrix factorization (sparseNMF), we estimate prototype color and density map of each stain in an unsupervised manner to perform stain separation. To color normalize a given source image, we combine its stain density maps with the stain color prototypes of a target image whose appearance was preferred by pathologists. In this way, the normalized image preserve the biological structure encoded in the stain density of the source image. Both the proposed sparseNMF stain separation and color-normalization techniques yield higher correlation with ground truth than the state of the art. They are also rated qualitatively higher than other techniques by a group of pathologists. We further propose a computationally faster extension of this technique for large whole-slide images that selects an appropriately small sample of patches to compute the color prototypes of each stain instead of using the entire image. The fast scheme achieves a 20-folds acceleration, which does not only greatly enhance the analysis efficiency, but also allow its clinical applications to become practically feasible.
|